“Water, water, every where, Nor any drop to drink.”
The Rime of the Ancient Mariner (1834 text)
by Samuel Taylor Coleridge
Kris T. Huang, MD, PhD, CTO
Deep learning requires data. Lots of it. There’s lots of medical data, almost 25 exabytes according to IEEE Big Data Initiatives [1], so where’s the problem? The problem is that more than 95% of medical data is unstructured, in the form of raw pixels (90%+) or text, essentially putting it out of reach of large scale analysis.

So what do you do when you don’t have that much?
Recently we released the last episode of our series exploring a few of the eccentricities of deep learning, this time about data augmentation. Once more playing the part of Deep Learning (DL), I zip through several cases in rapid succession while poor Machine Perception (MP) seems to lag behind, managing to get through only one case. Or so it seems.
If you look more closely at the case images, in reality DL had been looking at the same case, just rotated a bit differently each time. MP, though seemingly slower, progressed to a real and different second case image, but that doesn’t stop DL from gloating a bit about speed, even if it might be a while before reaching the actual second case.

Image credit: Bharath Raj (link)
This artificial data generation strategy is so-called “data augmentation.” Basically you turn one case into hundreds, thousands, or even millions of artificial ones. For example, because most convolutional networks do not recognize rotation, a common way to impart some degree of rotation invariance is to… rotate the image yourself and present it during training as a new and different case. But there’s the rub — even having “infinite” data does not mean infinite insight. Do the additional generated cat images above provide further insight to a human viewer about other cats, or even just the template cat on the left? Has the neural network really generalized the concept of rotation, or even the image content? What happens if the model is tested with an image rotated or translated beyond the trained range?
According to a paper earlier this year (2019) detailing Deep Learning Image Registration (DLIR), a framework for unsupervised deformable image registration [2], here’s what happens:
“The error is highly influenced by outliers, likely caused by the limited dataset size. Large initial landmark distances were scarcely available for training, which influenced registration performance…”
and later,
“…conventional image registration outperformed DLIR. Most likely caused by the amount of available representative training data. The employed augmentations were insufficient, and large deformations were not corrected by DLIR. Possibly by adding more training data results will improve.”
DIR-Lab, a 4D-CT dataset with manually generated ground truth points for maximum and minimum inhalation phases, consists of 10 cases with 10 phases per case for a total of 100 scans. Using leave-one-out cross-validation, there were only 810 permutations available for training. Since the maximum and minimum phases were used for validation, the use of leave-one-out cross-validation means that, by definition, DLIR was trained on phases with less extreme differences in respiratory volume. Inclusion of the max and min phases might improve the results, but nonetheless, it provides strong evidence that generalization is not occurring, considering that exceeding the training range by at most only about 10% produced inferior results.
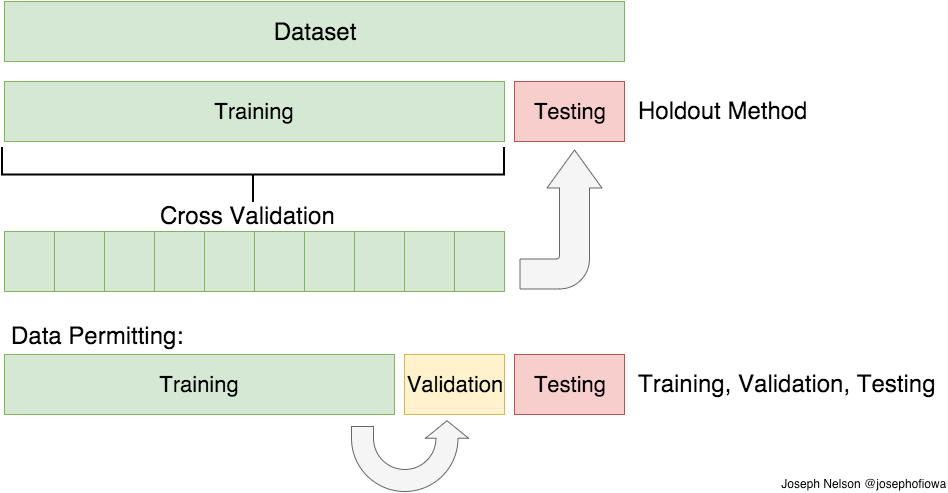
Image credit: https://medium.com/@josephofiowa
As for including the extremes of the DIR-Lab dataset, prior work with supervised deep learning showed that even providing the ground truth data, in addition to augmentation with random “large” transformations, still produced inferior results [3]. It’s likely that the low frequency of large displacements prevents “learning” or fitting these data points, essentially relegating them as outliers. For DLIR, attempts to improve fitting by stacking multiple sequential DLIR stages resulted in undesirable singularities and image folding.In other words, if you don’t have lots of data, augmenting it will not save the day. By extension, increasing the volume of data doesn’t necessarily increase insight, if it isn’t there in the first place.
As for the generalizability of all this, well, If you ever see a CT of the lungs in clinical practice that looks like this:
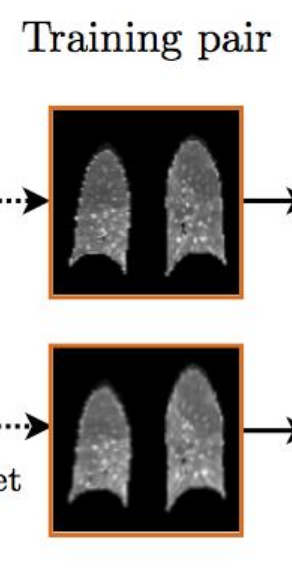
Image from [3].
image registration might be a lesser concern.
References
- A. Kuo, “Mining Health Big Data: Opportunities and Challenges,” presented at the Health 2.0 Asia 2016: South Korea and China, Nanjing, China, 10-Nov-2016.
- de Vos, B. D. et al. A deep learning framework for unsupervised affine and deformable image registration. Medical Image Analysis 52, 128–143 (2019).
- Eppenhof, K. A. . J., Lafarge, M. W., Moeskops, P., Veta, M. & Pluim, J. P. W. Deformable image registration using convolutional neural networks. in Medical Imaging 2018: Image Processing (eds. Angelini, E. D. & Landman, B. A.) 10574, 27 (SPIE, 2018).
